Integration example 3: Package for MAGs (Binning-level) with extdb¶
This tutorial will guide you through an integration of an example package, that requires an external database and it is referred to MAGs module. If this is your first time that reads a guide to our contribute section, we suggest you to read first the other two example.
What we need for this integration¶
Understand the Stream-level: in this case
Binning-basedModule name:
mags_func_annotationPackage name:
mags_kofam_scanKnow which are the conda dependencies for this packages, or the conda package name.
Know the code to actual integrate and execute the package.
Step 1: Clone/fork the repository and install Geomosaic¶
Since the final strategy is to make a pull request to the main repository, we suggest to fork our repo and then clone it (in the SSH way)
git@github.com:<YOURNAME>/Geomosaic.git
Install the Geomosaic conda environment. You can follow the Installation Guide.
Remember to replace <YOURNAME> with your GitHub user account.
Once you have cloned the repository, open the directory created with the clone and also create another branch specifying the name of the package that you are going to integrate
git checkout -b mags_kofam_scan
Step 2: Create the module folder (if does not exists)¶
Note
As we can intuitively think, KOfam Scan is a package for the functional annotation, in this case for the mags. Before this integration, the only package that belonged to this module was DRAM. However, DRAM usually takes in input a folder that contains all the fasta files of the mags, which is different from the input of the kofam scan, which takes the predicted orf from a single MAG.
Due to this difference, both packages cannot be in the same modules as the dependencies are different. So I decided to change the module belonging to DRAM, calling it mags_metabolism_annotation and insert mags_kofam_scan in the mags_functional_annotation, which depends on the mags_orf_prediction. Modules names are just to describe what the packages do, however, it would have been the same if it was mags_functional_annotation for DRAM and mags_functional_annotation_2 for kofam scan.
Step 3: Create the package folder¶
We need to create the package folder inside the corresponding module, which in this case is mags_func_annotation. Since we are going to integrate the program called KOfam Scan for MAGs, we can create a folder called mags_kofam_scan.
Warning
Do not use any special characters or insert spaces in the name.
Highlight
Just rely on underscore and all lower-case characters
Step 4: Create package’s snakefiles¶
Now we need to create the three files where we are going to implement all the necessary code:
Snakefile.smkSnakefile_target.smkparam.yaml
For now you can leave them empty.
Important
The names for this file are standard and are the same for each package. Do not change the filenames.
Step 5: create the corresponding conda env file¶
For this step, we don’t need to create the corresponding conda env file as we have already created for the Integration Example 2
Now we can write our code inside the Snakefile
Step 6: Link the module/package to the Geomosaic core¶
So in this section we need to link our new module and/or our new package to the already existing core of Geomosaic, which is represented by the file called gmpackages.json.
Let’s see how to do it.
Step 6.1: order section¶
Since this module depends on the predicted orfs from each mags, we insert this modules after the mags_orf_prediction.
...
"mags_orf_prediction",
"mags_domain_annotation",
"mags_func_annotation",
...
Step 6.2: graph section¶
Since this module depends on the predicted orfs from each mags, we insert this modules linked to the mags_orf_prediction.
...
["mags_orf_prediction", "mags_func_annotation"],
...
Step 6.3: modules section¶
In the corresponding modules section, we need to add the name of our package in our choices, which is a dictionary containing all the packages belonging to that module.
In particolar, the key (the blu string in the image) is the string that will come out in the terminal as a choice, during the workflow decision, while the value (in orange) is the actual name of the package, the one that we used also to create the folder in step 3.
Important
Package name on the value must match with the folder created in the step 3
Here we can see how DRAM was moved into the mags_metabolism_annotation module.
"modules": {
...
"mags_metabolism_annotation": {
"description": "Module: Perform annotation of metabolism on filtered MAGs [Binning-based]",
"choices": {
"DRAM on MAGs": "mags_dram"
}
},
...
"mags_func_annotation": {
"description": "Module: perform functional annotation on ORF retrieved from filtered MAGs [Binning-based]",
"choices": {
"KOfam Scan on MAGs": "mags_kofam_scan"
}
},
...
Step 6.4: additional_input section¶
If the package does require any additional input, you can integrate this input in the corresponding section of additional_input. In this case we don’t need to put any additional argument.
Highlight
Additional arguments are parameters that are widely known in the metagenomic workflow and that should be chosen by the user, as for example Completeness and Contamination.
In this section we have inserted also the possibility to specificy a folder that contains HMM models (for assembly_hmm_annotation and mags_hmm_annotation), as well as the name of the output folder these two modules in order to have different output name folder for different sets of HMMs.
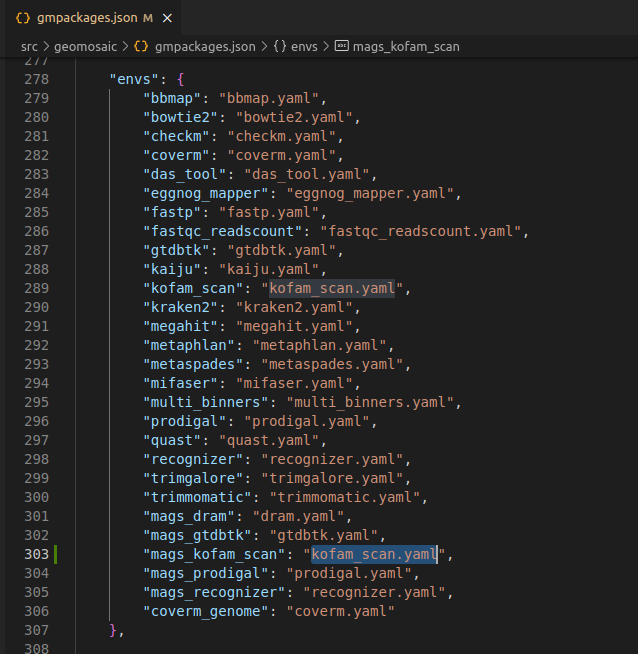
Step 6.5: envs section¶
This section is very simple, we only need to add the conda env file for our package that we have created in the Step 5. This filename must have the same package name. In this case kofam_scan.
Step 6.6: external_db section¶
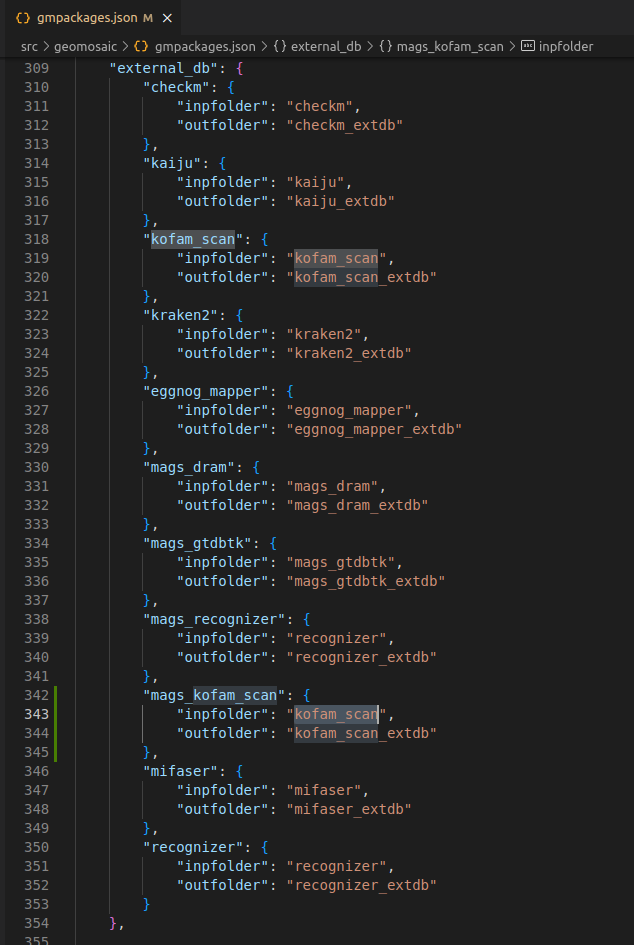
This section is useful to organize external databases for the package that we are going to integrate. In this example, we need an external database (extdb).
In this section:
each package that requires an extdb has a key which contains two other keys:
inpfolder: its value should be the name of the packageoutfolder: must be the name of the folder in which the external databases is going to be downloaded. The pattern is: the name of the package followed by the_extdbsuffix.
Highlight
In some cases, the same tool can be integrated at multiple levels, such as assembly-based and binning-based, as it was for the recognizer package and mags_recognizer. It is important to note that both integrations rely on the same external database. Therefore, in this section, we specify the same inpfolder and outfolder.
See also section 7.3 Bis in the Integation Example 2.
So in this section we are going to write as follows since the external database is the same for the KOfam Scan that we have integrated in the assembly_func_annotation (Integration Example 2).
Step 6.7: gathering section¶
This section is not mandatory. However it is useful if we want to compose some master tables or plots from the results of our package from all the samples computation. In this case, we are not interest in this section. However we could create a script that creates a table of all the reads count for each sample.
At the time of writing, this is the last section in the gmpackages.json.
Step 7: How to organize the code for the External Database (extdb).¶
You can see this section in the Integration example 2.
Step 8: Write the actual code.¶
To my knowledge, for the binning-based stream level there are two types of tool:
the first one are those that take into consideration as input all the fasta files inside a folder, like DRAM or GTDBTK. Indeed, their dependency is the
mags_retrieval.the second regards instead the tools that mainly take as input one fasta file at the time, like Prodigal or KOfam Scan which takes in input one predicted orf (fasta file) at the time.
For this latter case, it is essential to retrieve all the mags that we have and then refer to the fasta file for that mags. Many example are already integrated in Geomosaic, so we can start from that.
We can use the template of the mags_recognizer from the module mags_domain_annotation.
Step 8.1 Snakefile: input/output section¶
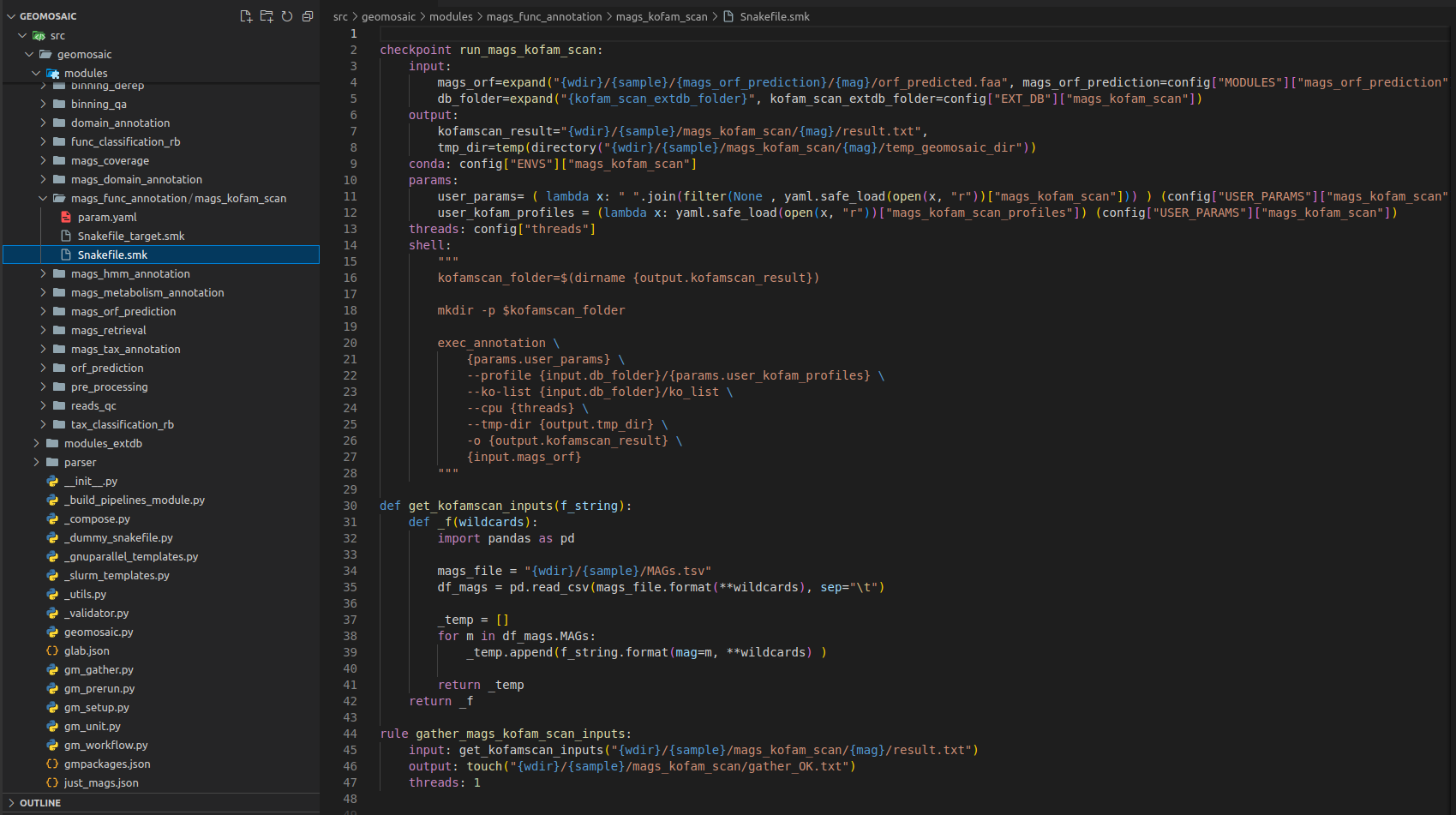
We need to change the rule definition with the package name, composed also of the prefix run_.
Our input section is fine, as we need only the predicted orf from each mags (
mags_orf_predictionmodule).In output section usually we put the folder output that must be the same of the package name. However if you know that your package is going to provide in output a specific file, you can even increase the detail of this section by inserting also that file.
Step 8.2 Snakefile: threads section¶
The threads section is fine like this. If we know that is not possible to execute our package through parallelization we can put in this section 1, otherwise we can leave it as it is.
Step 8.3 Snakefile: conda section¶
In this section, we only need to put our package name.
Step 8.4 Snakefile: params section¶
In each package we put at least a param variable called user_params, which is going to read the param.yaml file that we have created in the Step 4. The code to read user parameters, is almost always the same (so you don’t need to modify it):
user_params=( lambda x: " ".join(filter(None , yaml.safe_load(open(x, "r"))["mags_kofam_scan"])) ) (config["USER_PARAMS"]["mags_kofam_scan"])
Just replace mags_kofam_scan with your package name.
Since this package accept a profile database for the annotation, we have inserted another param called mags_kofam_scan_profiles (Section 10) that is read by the following line
user_kofam_profiles = (lambda x: yaml.safe_load(open(x, "r"))["mags_kofam_scan_profiles"]) (config["USER_PARAMS"]["mags_kofam_scan"])
Step 8.5 Snakefile: shell section¶
This is the section in which we are going to put the actual code to execute our programs.
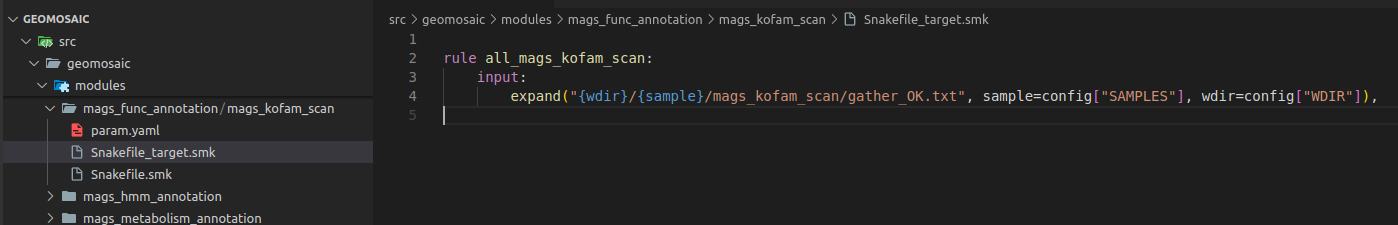
Step 9: Snakefile Target¶
In our file Snakefile_target.smk we only need to write few rows. First, the name of the rule must be the same name of the package name with the all_ prefix. And then we need to change the rows in the input section, and we need to specify the same folder output as in this case was our only output that we specified in the Snakefile.smk.
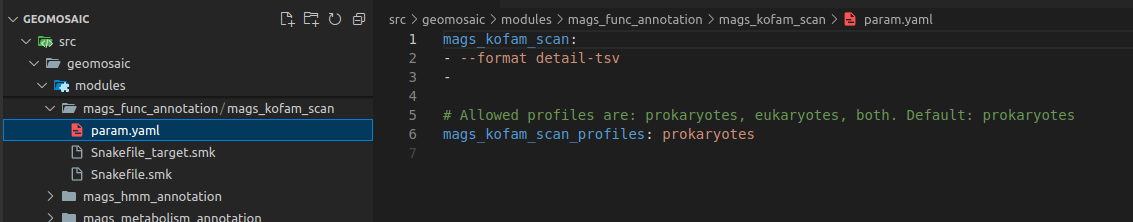
Step 10: Param.yaml file¶
The param.yaml is a file in which the user, before the execution of the workflow, can insert all the optional parameters belonging to the package as bullet points. In this case, we only need to open this file and add the following lines:
Test the integration¶
Now we should test the integrated package. Activate the conda environment of geomosaic. Updated geomosaic by doing
pip install .
Once we have tested, we can commit the changes and create the pull request.