geomosaic workflow¶
Caution
Before running a workflow from scratch up to the modules related to MAGs annotation, please read this suggestion.
Overview¶
After having successfully executed the setup command, Geomosaic is ready to create your desired workflow.
geomosaic workflow --help
usage: geomosaic workflow -s SETUP_FILE [-t THREADS] [-p {just_mags,glab,None}] [-m MODULE] [-h]
DESCRIPTION: It allows to choose the desired modules and the relative packages. Based on you choices, the command will create a Snakefile (in the geomosaic directory) with the chosen modules, the config file for snakemake, and a graph image to show the created workflow
Required Arguments:
-s SETUP_FILE, --setup_file SETUP_FILE
Geomosaic setup file created from the geomosaic setup ... command.
Optional Arguments:
-t THREADS, --threads THREADS
Threads to use (per sample).
-p {just_mags,glab,None}, --pipeline {just_mags,glab,None}
Execute a default Giovannelli's Lab pipeline of Geomosaic (Completeness 50, Contamination 10). The pipeline 'glab' is a full
pipeline without the two modules on HMM annotation (assemby and mags). The pipeline 'just_mags' is a minimal set of modules to
retrieve MAGs without any annotation (Completeness: 50, Contamination: 10).
-m MODULE, --module_start MODULE
Module where to start creating the workflow (Default: pre_processing)
Available Modules:
- pre_processing - Module: Pre processing with quality check
- reads_qc - Module: Quality check (and/or Reads Count) of the reads after Pre-Processing
- func_classification_rb - Module: Functional classification [Read-based]
- tax_classification_rb - Module: Taxonomic classification [Read-based]
- assembly - Module: Assembly
- assembly_func_annotation - Module: Functional Annotation on Assembly [Assembly-based]
- assembly_qc - Module: Assembly quality evaluation with metrics [Assembly-based]
- assembly_readmapping - Module: Read mapping the processed reads to the Assembly [Assembly-based]
- assembly_coverage - Module: Assembly coverage computation [Assembly-based]
- assembly_tax_annotation - Module: Taxonomic Annotation on Assembly [Assembly-based]
- orf_prediction - Module: perform ORF prediction [Assembly-based]
- domain_annotation - Module: Domain annotation on Assembly [Assembly-based]
- assembly_hmm_annotation - Module: tracking coverage for genes of interest through HMM models [Assembly-based]
- assembly_orf_annotation - Module: Functional Annotation on Assembly (ORFs) [Assembly-based]
- binning - Module: Binning
- binning_derep - Module: Binning Deeplication [Binning-based]
- binning_qa - Module: Binning Quality Evaluation [Binning-based]
- mags_retrieval - Module: get MAGs based on Contamination and Completeness threshold [Binning-based]
- mags_func_annotation - Module: perform functional annotation on MAGs [Binning-based]
- mags_tax_annotation - Module: Perform taxonomic annotation of filtered MAGs [Binning-based]
- mags_orf_prediction - Module: perform ORF prediction for each retrieved MAG [Binning-based]
- mags_domain_annotation - Module: perform domain-based annotation for each retrieved MAG [Binning-based]
- mags_orf_annotation - Module: perform functional annotation on ORF retrieved from filtered MAGs [Binning-based]
- mags_coverage - Module: Computing read coverage for each retrieved MAG [Binning-based]
- mags_hmm_annotation - Module: tracking coverage for genes of interest through HMM models on MAGs [Binning-based]
Help Arguments:
-h, --help show this help message and exit
Arguments¶
This command has both one required and different optional arguments:
REQUIRED
(
-s) Specifiy the name of the Geomosaic config file, obtained with thesetupcommand.
OPTIONAL
(
-p <pipeline>) With this flag, the user can choose already pre-defined workflows. Currently, two pipeline are integrated:glabwhich contains most of the annotation in all the stream levels,just_magsthat allows to run the minimal modules to obtain the mags. Both pipelines are now though to run withcompletenessandcontaminationvalues of 50 and 10 respectively.(
-m) Module where to start creating the workflow (Default: pre_processing)(
-t) Threads to use (per sample)
What to expect from this command¶
After completing this command, Geomosaic generates three files in its working directory:
Snakefile.smk- the Snakefile with the rules for each chosen module. This is the real pipeline;config.yaml- the config file for snakemake execution (which is different from the config file generated bygeomosaic setup);Snakefile_extdb.smk- An additional Snakefile containing all the codes to setup external databases of the chosen packages. This snakefile will be the first to be executed, in order to download the required data.
It’s all automated.
Example usage geomosaic workflow¶
IMPORTANT: the following images may not refer to the modules that are integrated in the current version of Geomosaic. However, these images are very useful to understand how it works the creation of the workflow works.
By executing this command, Geomosaic starts loading the provided config file, and then it asks for the desired package to be used for each module. It is important to considered the above mentioned dependencies among the modules:
geomosaic workflow -s gmsetup_exp2023.yaml
Geomosaic Process: Loading variables from Geomosaic setup file...
--> OK <--
[PRE_PROCESSING] - Module: Pre processing with quality check
0) -- Ignore this module (and all successors) --
1) fastp
2) Trim-Galore
3) Trimmomatic
1
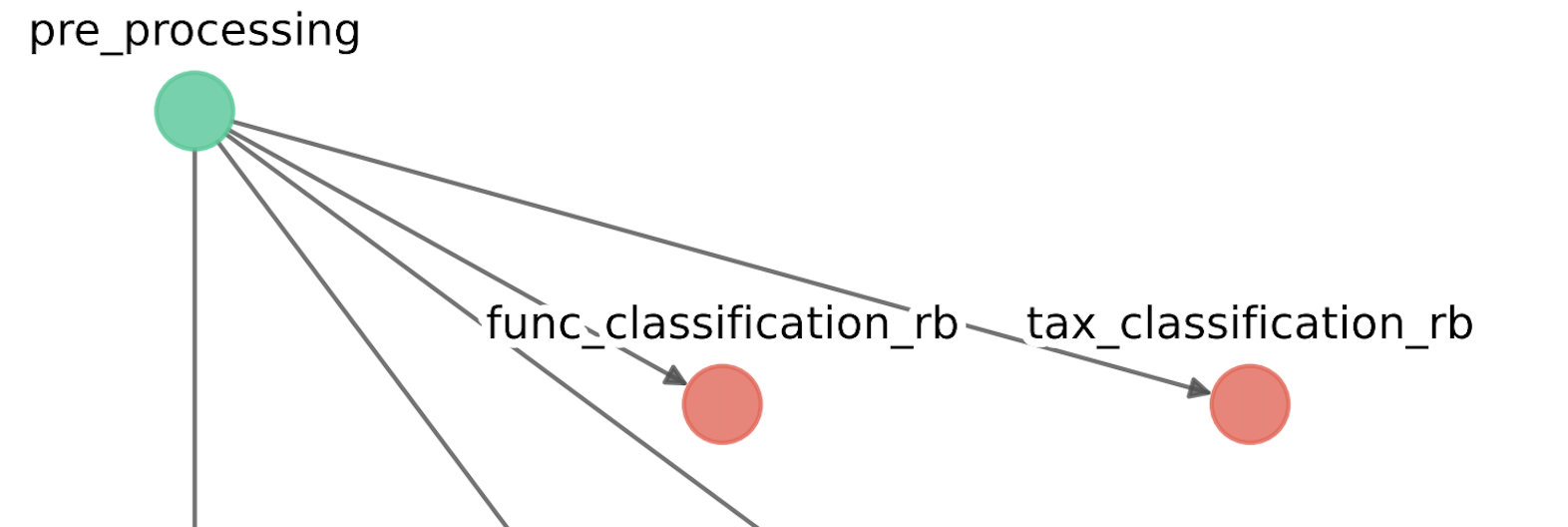
[FUNC_CLASSIFICATION_RB] - Module: Functional classification [Read-based]
0) -- Ignore this module (and all successors) --
1) mi-faser (GS-21-all)
0
[TAX_CLASSIFICATION_RB] - Module: Taxonomic classification [Read-based]
0) -- Ignore this module (and all successors) --
1) Kaiju
2) MetaPhlAn
0
As it is shown, geomosaic will ask for each module the desired package, waiting for the user input.
this input must be a integer value corresponding to the choice. Typing 0 will ignore the module and all the next modules that depends on it. In this case, fastp was chosen by typing 1, and the modules func_classification_rb tax_classification_rb were ignored.
[ASSEMBLY] - Module: Assembly
0) -- Ignore this module (and all successors) --
1) MetaSpades
2) MegaHit
1
[ASSEMBLY_READMAPPING] - Module: Assembly evaluation by read mapping [Assembly-based]
0) -- Ignore this module (and all successors) --
1) Bowtie2
2) BBMap
1
[ASSEMBLY_QC] - Module: Assembly evaluation with metrics [Assembly-based]
0) -- Ignore this module (and all successors) --
1) Quast
0
[ASSEMBLY_TAX_ANNOTATION] - Module: Taxonomic Annotation on Assembly [Assembly-based]
0) -- Ignore this module (and all successors) --
1) Kraken2
1
[ORF_PREDICTION] - Module: perform ORF prediction [Assembly-based]
0) -- Ignore this module (and all successors) --
1) Prodigal
1
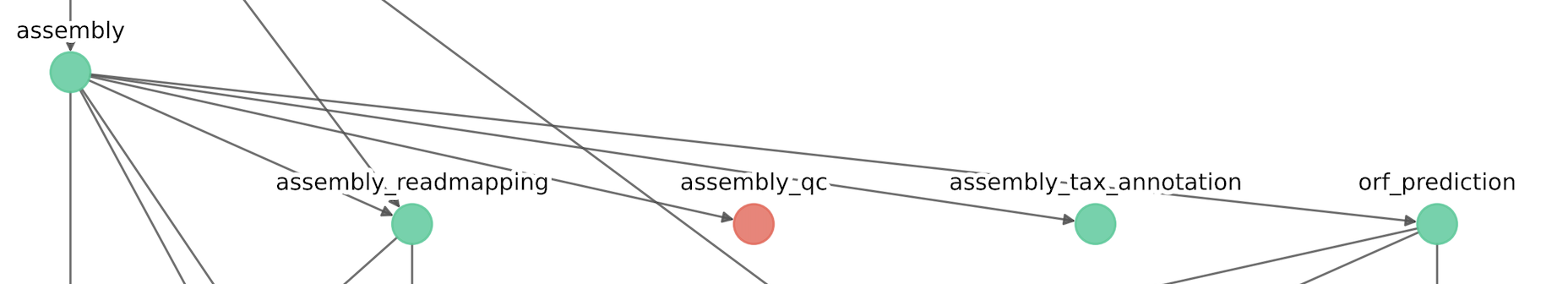
[ASSEMBLY_COVERAGE] - Module: Assembly coverage computation [Assembly-based]
0) -- Ignore this module (and all successors) --
1) CoverM (Contigs)
0
[ASSEMBLY_FUNC_ANNOTATION] - Module: Functional Annotation on Assembly [Assembly-based]
0) -- Ignore this module (and all successors) --
1) eggNOG-Mapper
1
[DOMAIN_ANNOTATION] - Module: Domain annotation on Assembly [Assembly-based]
0) -- Ignore this module (and all successors) --
1) reCOGnizer
0
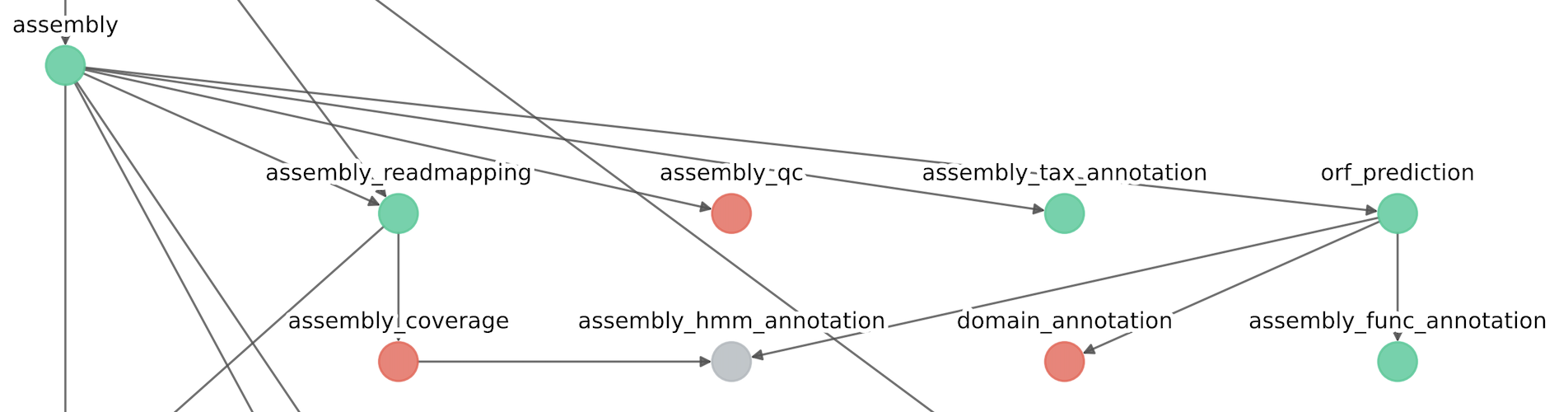
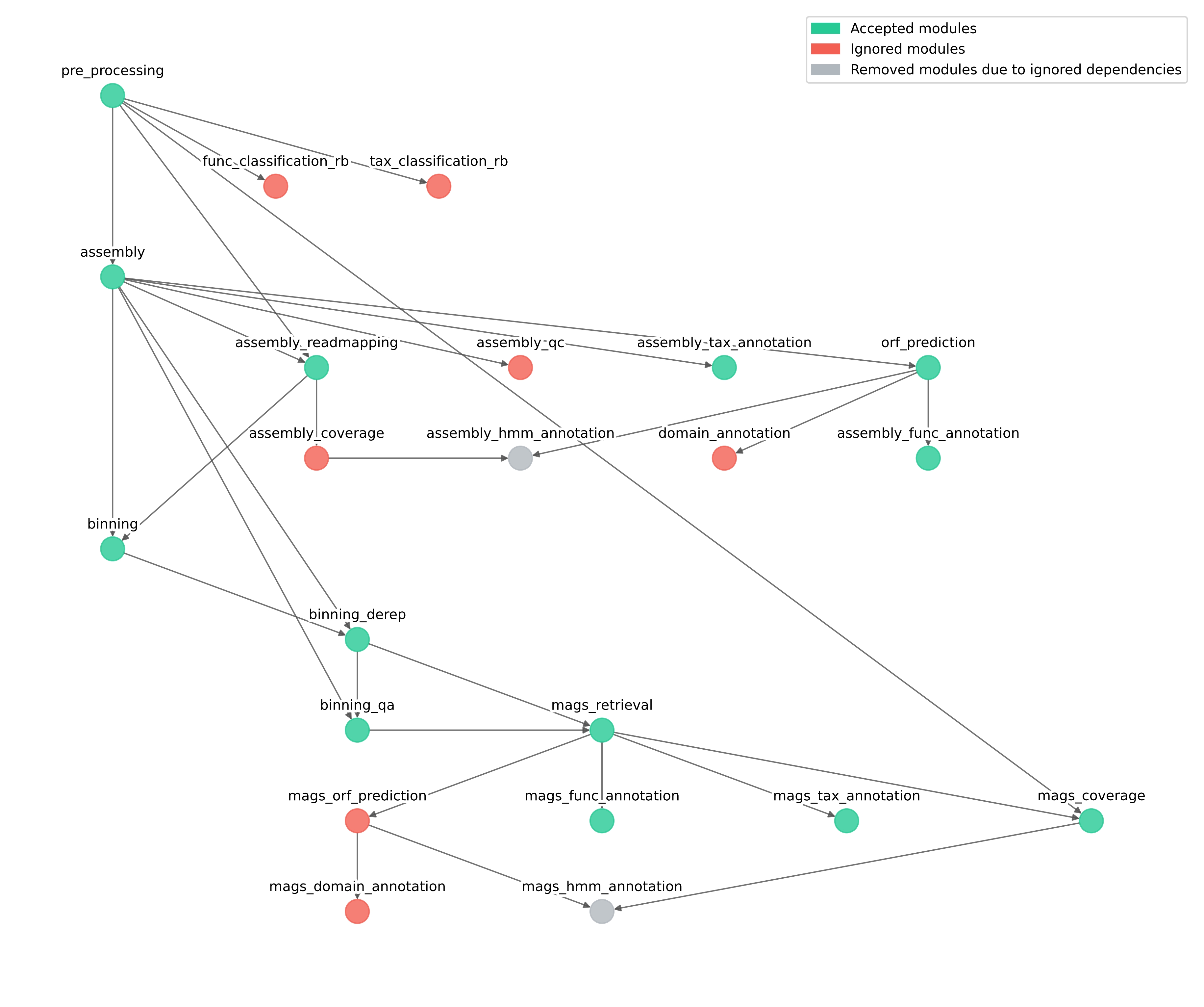
IMPORTANT: Since that the module assembly_coverage is ignored, the module assembly_hmm_annotation is skipped as it depends upon assembly_coverage.
At this point Geomosaic continues asking for modules
[BINNING] - Module: Binning
0) -- Ignore this module (and all successors) --
1) MaxBin2 + MetaBat2 + SemiBin2
1
[BINNING_DEREP] - Module: Binning Deeplication [Binning-based]
0) -- Ignore this module (and all successors) --
1) DAS_tool
1
[BINNING_QA] - Module: Binning Quality Evaluation [Binning-based]
0) -- Ignore this module (and all successors) --
1) CheckM
1
[MAGS_RETRIEVAL] - Module: get MAGs based on Contamination and Completeness threshold [Binning-based]
0) -- Ignore this module (and all successors) --
1) MAGs HQ
1
[MAGS_FUNC_ANNOTATION] - Module: Perform annotation of filtered MAGs [Binning-based]
0) -- Ignore this module (and all successors) --
1) DRAM on MAGs
1
[MAGS_TAX_ANNOTATION] - Module: Perform taxonomic annotation of filtered MAGs [Binning-based]
0) -- Ignore this module (and all successors) --
1) GTDBTK on MAGs
1
[MAGS_ORF_PREDICTION] - Module: perform ORF prediction for each retrieved MAG [Binning-based]
0) -- Ignore this module (and all successors) --
1) Prodigal on MAGs
0
[MAGS_COVERAGE] - Module: Computing read coverage for each retrieved MAG [Binning-based]
0) -- Ignore this module (and all successors) --
1) CoverM (Genome)
1
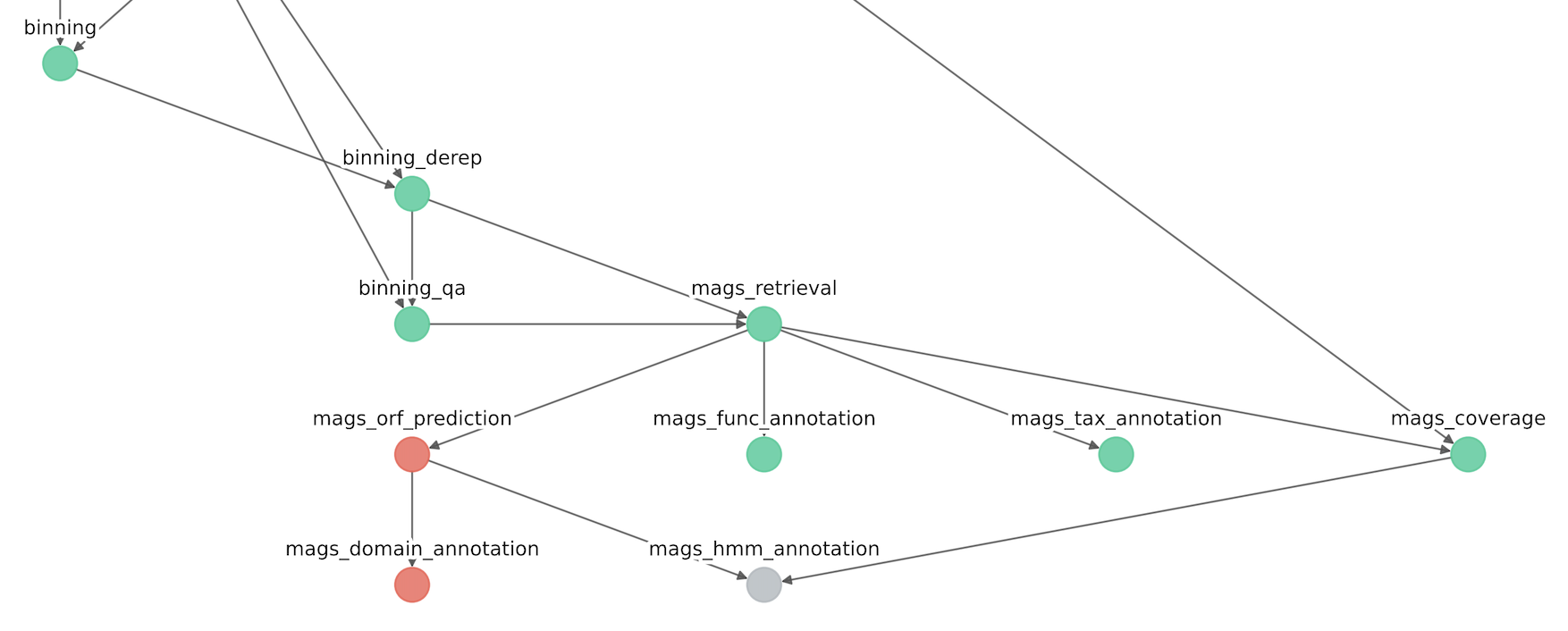
Also in this situation, since that the module mags_orf_prediction has been ignored, the module mags_hmm_annoation has been removed due to the ignored dependency.
After having chosen all desired modules, Geomosaic asks for values for additional parameters. For instance, in the workflow described above the module MAGs Retrieval was chosen. Since it regards the retrieval of high quality MAGs based on the contamination and completness, Geomosaic asks the user additional parameters.
[Additional Parameter to retrieve HQ MAGs] Description: COMPLETENESS parameters to filter retrieved bins
Required parameter: integer value - Suggested value: 90:
90
[Additional Parameter to retrieve HQ MAGs] Description: CONTAMINATION parameters to filter retrieved bins
Required parameter: integer value - Suggested value: 10:
10
Geomosaic Process: Building preliminary workflow to prepare all the database of your workflow...
--> OK <--
The resulting conceptual map of the modules should be as follows: