geomosaic setup¶
Overview¶
This commands should be the first one to be executed as it sets up the Geomosaic working directory with the corresponding folder and sample names.
geomosaic setup --help
usage: geomosaic setup -d DIRECTORY -t SAMPLE_TABLE [-s SETUP_FILE] [-c CONDAENV_GMFOLDER] [-e EXTERNALDB_GMFOLDER] [-u USERPARAMS_GMFOLDER]
[-f {tsv,csv,excel}] [-w WORKING_DIR] [-n PROJECT_NAME] [--move_and_rename] [--skip_checks] [-h]
DESCRIPTION: It creates the geomosaic working directory and the relative samples folders based on the provided sample table
Required Arguments:
-d DIRECTORY, --directory DIRECTORY
Path to the directory containing raw reads (fastq.gz files) (default: None)
-t SAMPLE_TABLE, --sample_table SAMPLE_TABLE
Path to the user sample table (default: None)
Optional Arguments:
-s SETUP_FILE, --setup_file SETUP_FILE
Output name for the geomosaic setup file (yaml extension). This file is necessary for the < geomosaic workflow > command. (default:
gmsetup.yaml)
-c CONDAENV_GMFOLDER, --condaenv_gmfolder CONDAENV_GMFOLDER
This option allows to provide a path folder in which geomosaic is going to install all the conda environments of your workflow. This
option is very useful if you want to execute Geomosaic for different set of reads, as here you can provide the same folder and
prevent multiple installation of the same conda environments. If not specified geomosaic will create a folder called 'gm_conda_envs'
inside the directory provided by the '-w' option. (default: None)
-e EXTERNALDB_GMFOLDER, --externaldb_gmfolder EXTERNALDB_GMFOLDER
This option allows to provide a path folder in which geomosaic is going to download all the external databases used by the packages
of your workflow. This option is very useful if you want to execute Geomosaic for different set of reads, as here you can provide
the same folder and prevent multiple donwload of the same external databases. If not specified geomosaic will create a folder called
'gm_external_db' inside the directory provided by the '-w' option. (default: None)
-u USERPARAMS_GMFOLDER, --userparams_gmfolder USERPARAMS_GMFOLDER
This option allows to provide a path folder in which geomosaic is going to put file for additional options/parameters that can be
used by the packages of your workflow. This option is very useful if you want to execute Geomosaic for different set of reads and
using the tools with the same sets of options/parameters of previous Geomosaic executions. If not specified geomosaic will create a
folder called 'gm_user_parameters' inside the directory provided by the '-w' option. (default: None)
-f {tsv,csv,excel}, --format_table {tsv,csv,excel}
Format of the provided table. Allowed: tsv, csv, excel (default: tsv)
-w WORKING_DIR, --working_dir WORKING_DIR
Is the Geomosaic working directory that has to be created for the pipeline execution. Default: 'geomosaic' folder created in the
current directory (default: geomosaic)
-n PROJECT_NAME, --project_name PROJECT_NAME
Name of the project. The first 8 Characters will be used for SLURM job name (default: Geomosaic_Workflow)
--move_and_rename Suggested flag if the provided raw reads directory is already a backup of the original files. In this case, geomosaic will create
only symbolic link of raw reads to its working directory. Note: This flag cannot be used if there are multiple files for each R1 and
R2 sample reads, as geomosaic will 'cat' them to a single file. (default: False)
--skip_checks If you are sure that every file is in its correct location and the sample names are filled correctly, you can skip checks with this
flags. However we do not suggest to use it. (default: False)
Help Arguments:
-h, --help show this help message and exit
Arguments¶
setup command has two required arguments:
REQUIRED
(
-t) the sample table including all names of the reads files and the desired sample names. It is expected to provide a table with three columns having the following headers:r1,r2,sample. For instance:r1
r2
sample
S1_L001_R1_fastq.gz
S1_L001_R2_fastq.gz
sample1
S1_L002_R1_fastq.gz
S1_L002_R2_fastq.gz
sample1
S2_L001_R1_fastq.gz
S2_L001_R2_fastq.gz
sample2
S2_L002_R1_fastq.gz
S2_L002_R2_fastq.gz
sample2
S3_L001_R1_fastq.gz
S3_L001_R2_fastq.gz
sample3
…
…
…
NOTE 1: Sample and reads name with no spaces: the values on thesamplecolumn (but also on ther1andr2columns) should not present any space as it increases the folder organization complexity. However, Geomosaic will perform a check (assertion) and possibly print an Error message describing the line that containes the space.NOTE 2: Different lines with the same sample name are allowed: sequencing data are often splitted for a single sample. In this case, is it quite common tocatthem all in one file; this is performed for both R1 (read R1) and R2 (read R2). Geomosaic allows the same sample name in multiple lines of the provided table, as it willcatall the reads R1 in one file R1 and all the reads R2 in one file R2. This case is also presented in the example table above (line 2,3 and 3, 4).In the
NOTE 2 scenario, Geomosaic will perform agroup byof R1 and R2 files based on thesamplecolumn to then use thecatcommand. For instance, by performing a group by on the provided example table, the following files are obtained:r1
r2
sample
[S1_L001_R1.fastq.gz, S1_L002_R1.fastq.gz]
[S1_L001_R2.fastq.gz, S1_L002_R2.fastq.gz]
sample1
[S2_L001_R1.fastq.gz, S2_L002_R1.fastq.gz]
[S2_L001_R2.fastq.gz, S2_L002_R2.fastq.gz]
sample2
[S3_L001_R1.fastq.gz, … ]
[S3_L001_R2.fastq.gz, …]
sample3
…
…
…
(
-d) the directory containing all raw reads. The name of the listed files has to match the ones provided in the tabular table.
The setup command has various optional arguments:
OPTIONAL
(
-s) The output name for the setup file of Geomosaic. Default:gmsetup.yaml(in the current directory).(
-c) This option is very useful if you are going to run geomosaic for different set of raw reads. By specifying this folder, geomosaic will not reinstall all the conda environments. Here you can provide the same folder and prevent multiple installation of the same conda environments. If not specified geomosaic will create a folder calledgm_conda_envsinside the directory provided by the-woption.(
-e) Similarly to the-cparameter, this option is very useful if you are going to run geomosaic for different set of raw reads. By specifying this folder, geomosaic can use already downloaded external databases. If not specified geomosaic will create a folder calledgm_external_dbinside the directory provided by the-woption.For example, let’s assume that in the following path
/mnt/storage/geomosaic_extdbare already presents the following databaseskaiju_extdb,kraken2_extdbandcheckm_extdb(that were downloaded with Geomosaic) are already present; you can use this option as-e /mnt/storage/geomosaic_extdb.
(
-u) Similarly to the-cand-eflags, this options allows you to specify the same folder path that you may have used in previous Geomosaic execution for theuser parameter folder. This is useful to execute same tools with the same options that you may have added in those executions. If not specified geomosaic will create a folder calledgm_user_parametersinside the directory provided by the-woption.(
-f) With this option you can specify the format separated value of the table provided with the flag-t.(
-w) The Geomosaic working directory to create for its execution. Default:geomosaicfolder created in the current directory.(
-n) A project name can be specified with this parameter. It is recommended to use a name without any space.(
--move_and_rename) You can use this option to directly move the reads file from the directory specified in-dparameter instead of creating another copy.NOTE 3: --move_and_rename parameter not allowed in NOTE 2 scenario: due to the need tocatall sample sequencing reads in one file, the--move_and_renameparameter is not allowed in theNOTE 2scenario. Indeed,--move_and_renameparameter can be used when there is a single file for both R1 and R2 sequencing reads.
What to expect from this command¶
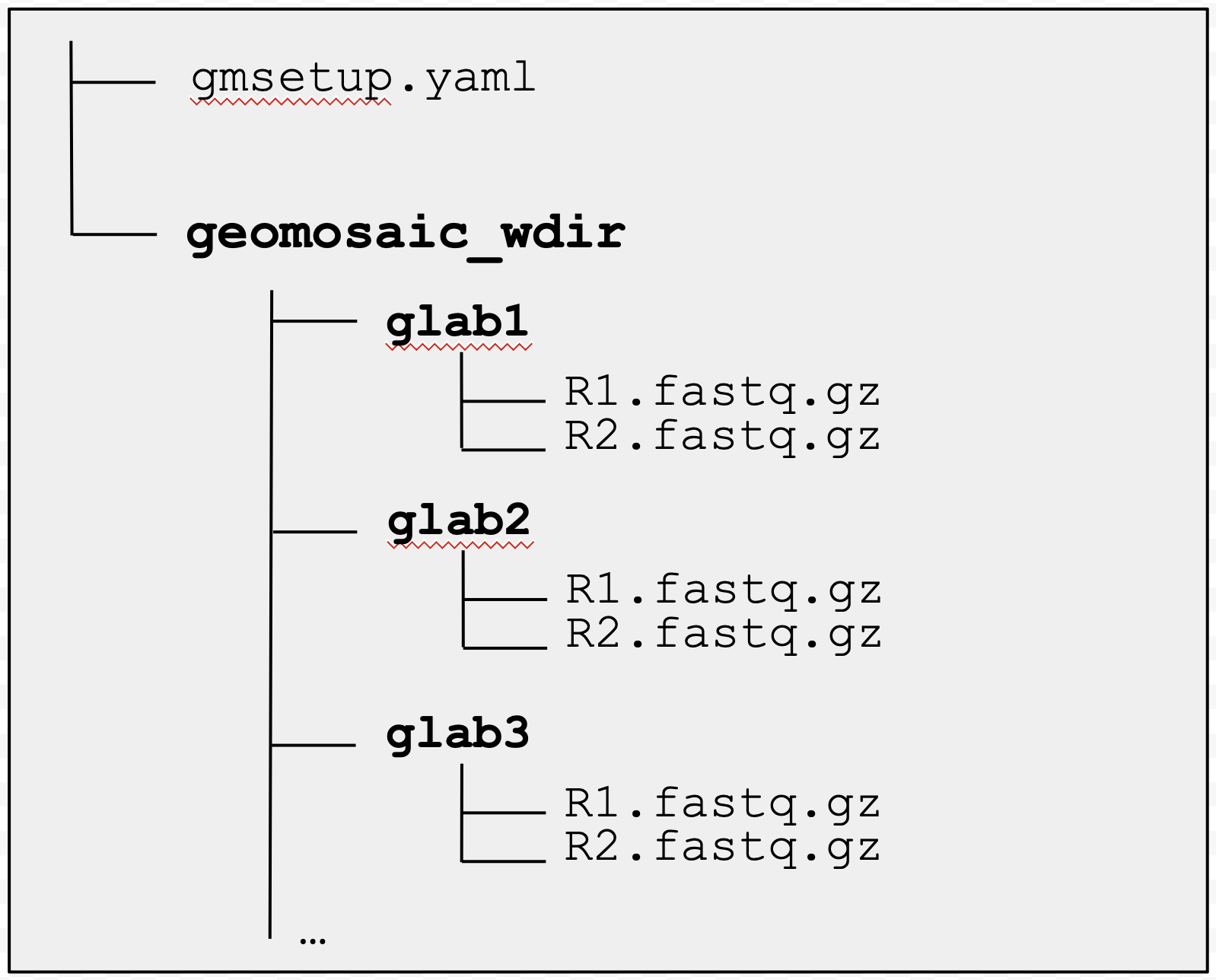
After completing this command you should see:
a geomosaic setup file, by default called
gmsetup.yamlthat contains some essential information for the next commands. Do not modify this file.and the working directory for geomosaic (eventually provided with the option
-w) which stores a directory for each of the user unique samples. Each sample folder contains containing two files:R1.fastq.gzR2.fastq.gz
Example usage geomosaic setup¶
Assuming my sequencing reads are in the folder termed sequencing_mg_expedition2023 and my tabular file is called sample_table_expedition2023.tsv,
the setup command can be executed as follows
geomosaic setup -d sequencing_mg_expedition2023 -t sample_table_expedition2023.tsv
or providing optional parameters to add further information
geomosaic setup -d sequencing_mg_expedition2023 \
-t sample_table_expedition2023.tsv \
-s gmsetup_exp2023.yaml \
-w ./geomosaic_exp2023 \
-n "Geomosaic EXP2023" \
--move_and_rename